PLUGINS for LIFE SCIENCE, GENOMICS, AND HEALTHCARE
Tools that make life science data searchable, traceable, and actionable.
Diskover’s Life Science plugin suite is designed to extract the signals researchers care about—sample IDs, runs, cases, projects, ownership, and status—then apply that context consistently across storage. The result is faster search, cleaner handoffs, and automation you can trust, without changing how teams store or access data..
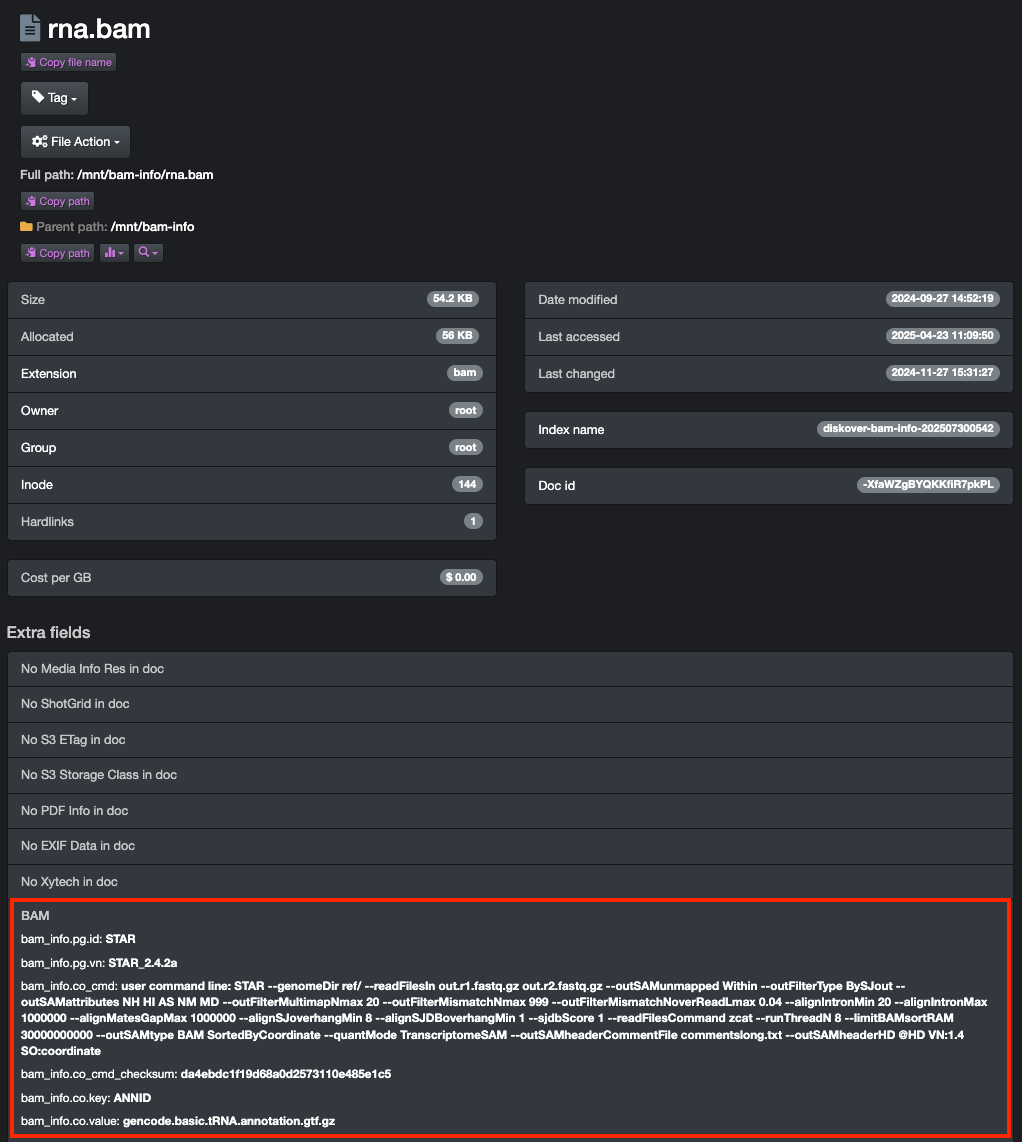
BAM plugin.
Optimize every data pipeline in your genomics ecosystem.
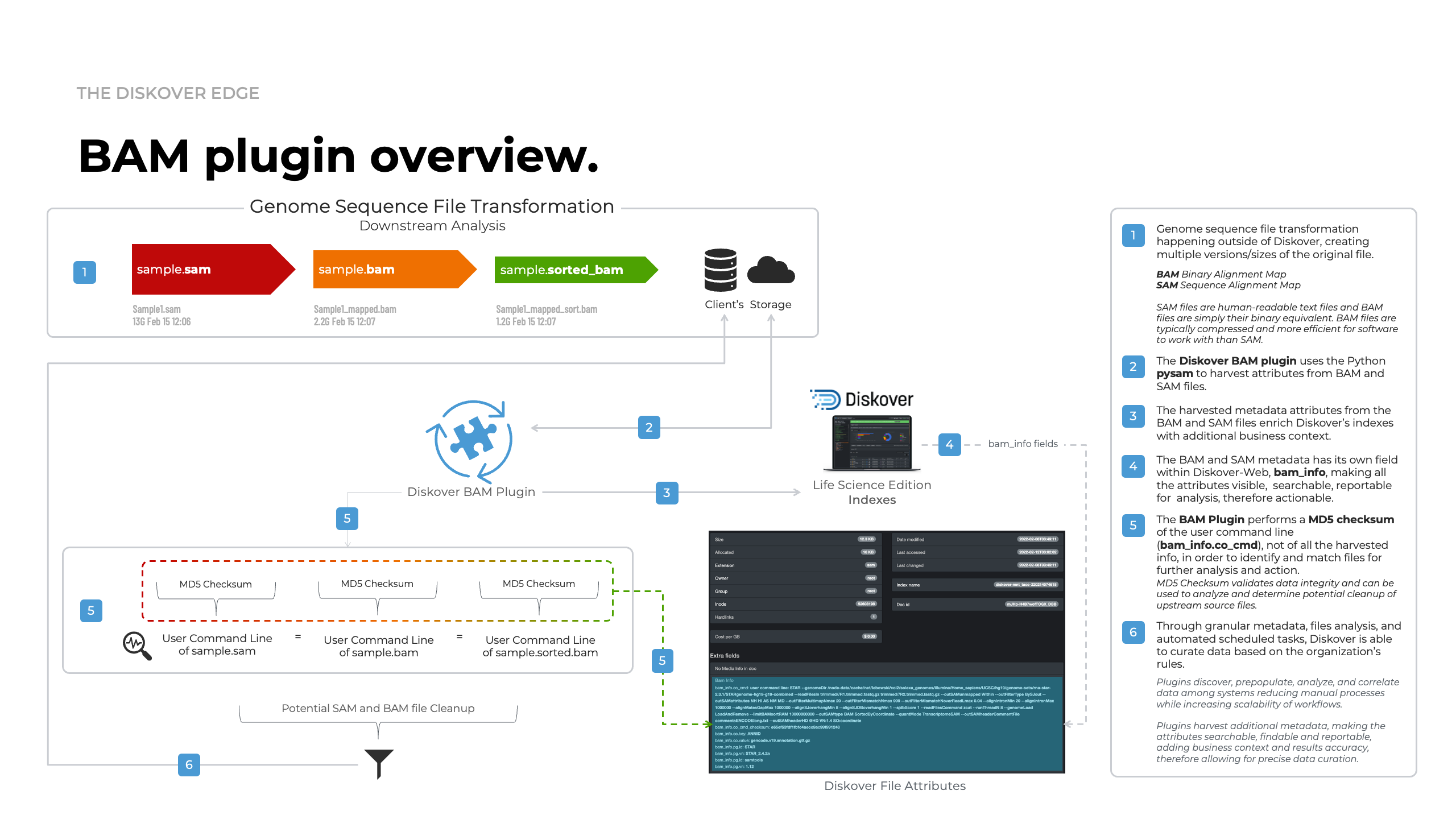
The BAM/SAM metadata Eerichment plugin extends Diskover’s indexing capabilities into bioinformatics pipelines by harvesting alignment and command-line metadata directly from BAM and SAM files—without requiring any read/write access, ensuring full data integrity.
How it works.
Why it matters.
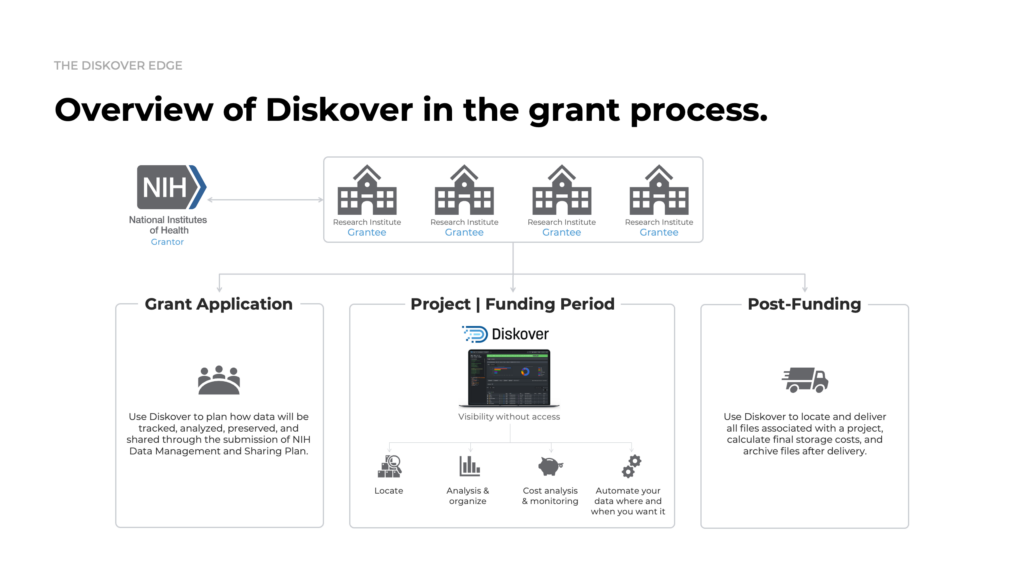
GRANT plugin.
Link datasets, grants, and research outcomes.
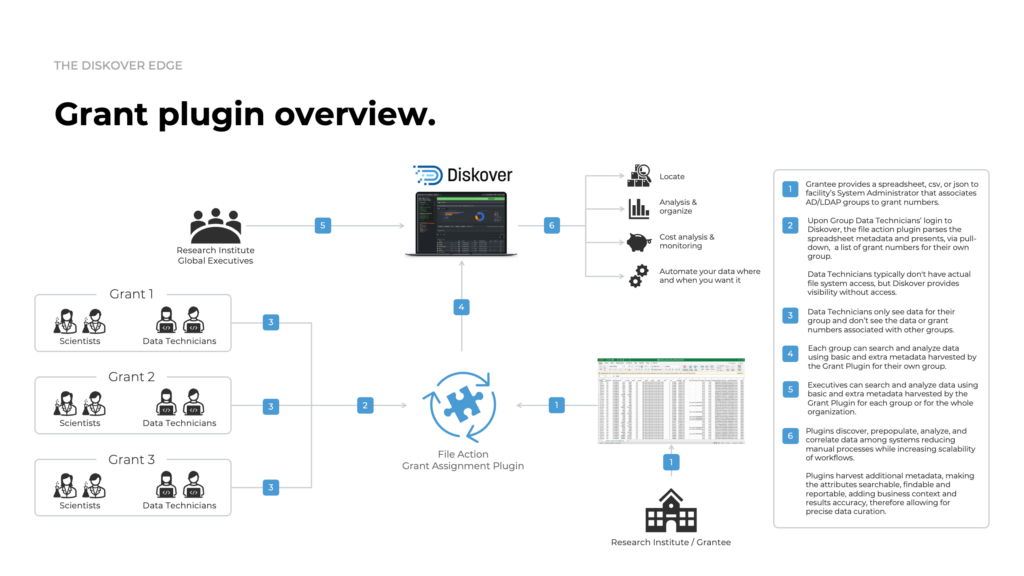
The Grant plugin brings transparency and intelligence to scientific data management by linking research datasets to grant metadata—enabling realiable compliance with NIH policies, accurate cost tracking, and automated lifecycle oversight.
How it works.
Why it matters.
Ready to bring order to your unstructured world?